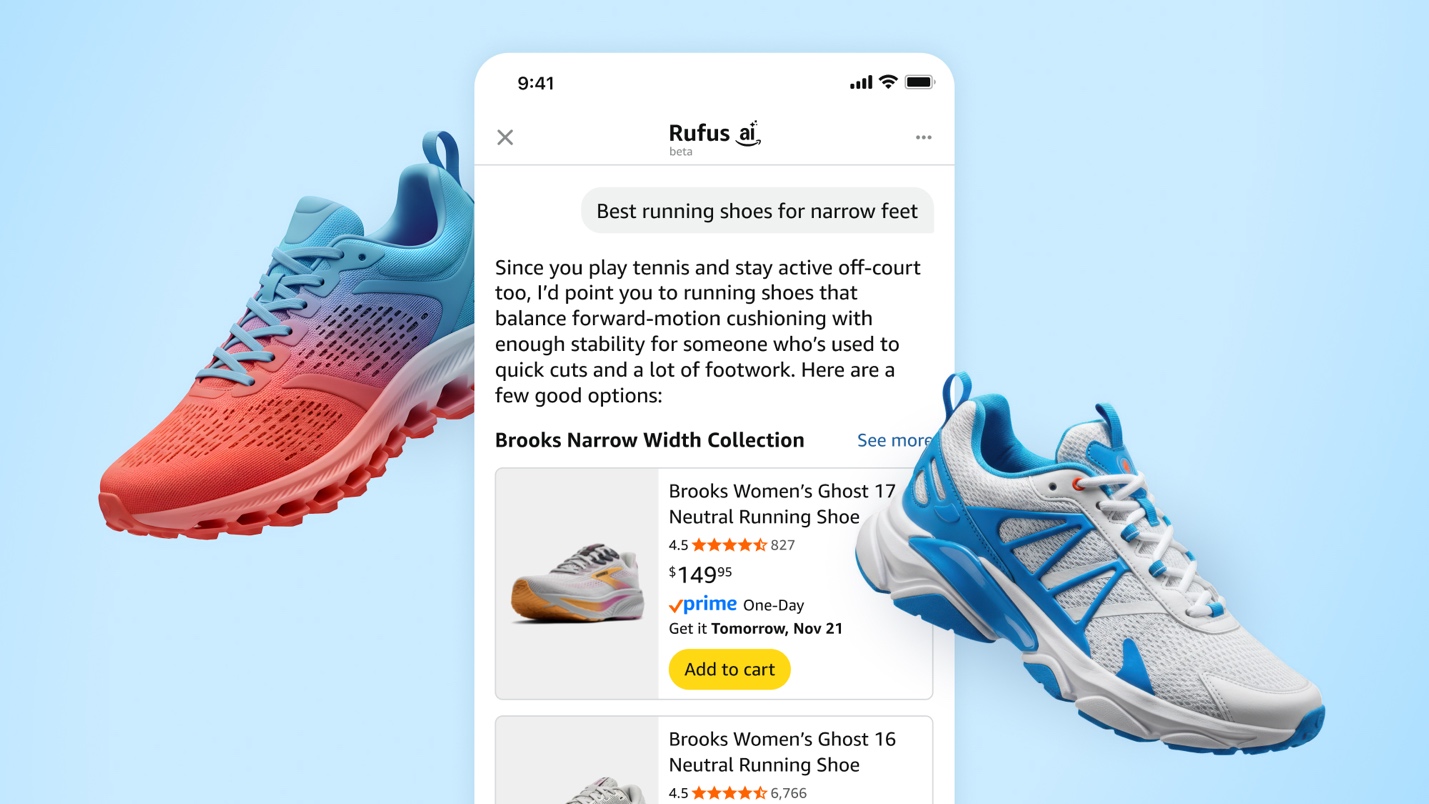
How Rufus scales conversational shopping experiences to millions of Amazon customers with Amazon BedrockArtificial Intelligence Our team at Amazon builds Rufus, an AI-powered shopping assistant which delivers intelligent, conversational experiences to delight our customers. More than 250 million customers have used Rufus this year. Monthly users are up 140% YoY and interactions are up 210% YoY. Additionally, customers that use Rufus during a shopping journey are 60% more likely to
Our team at Amazon builds Rufus, an AI-powered shopping assistant which delivers intelligent, conversational experiences to delight our customers. More than 250 million customers have used Rufus this year. Monthly users are up 140% YoY and interactions are up 210% YoY. Additionally, customers that use Rufus during a shopping journey are 60% more likely to Read More
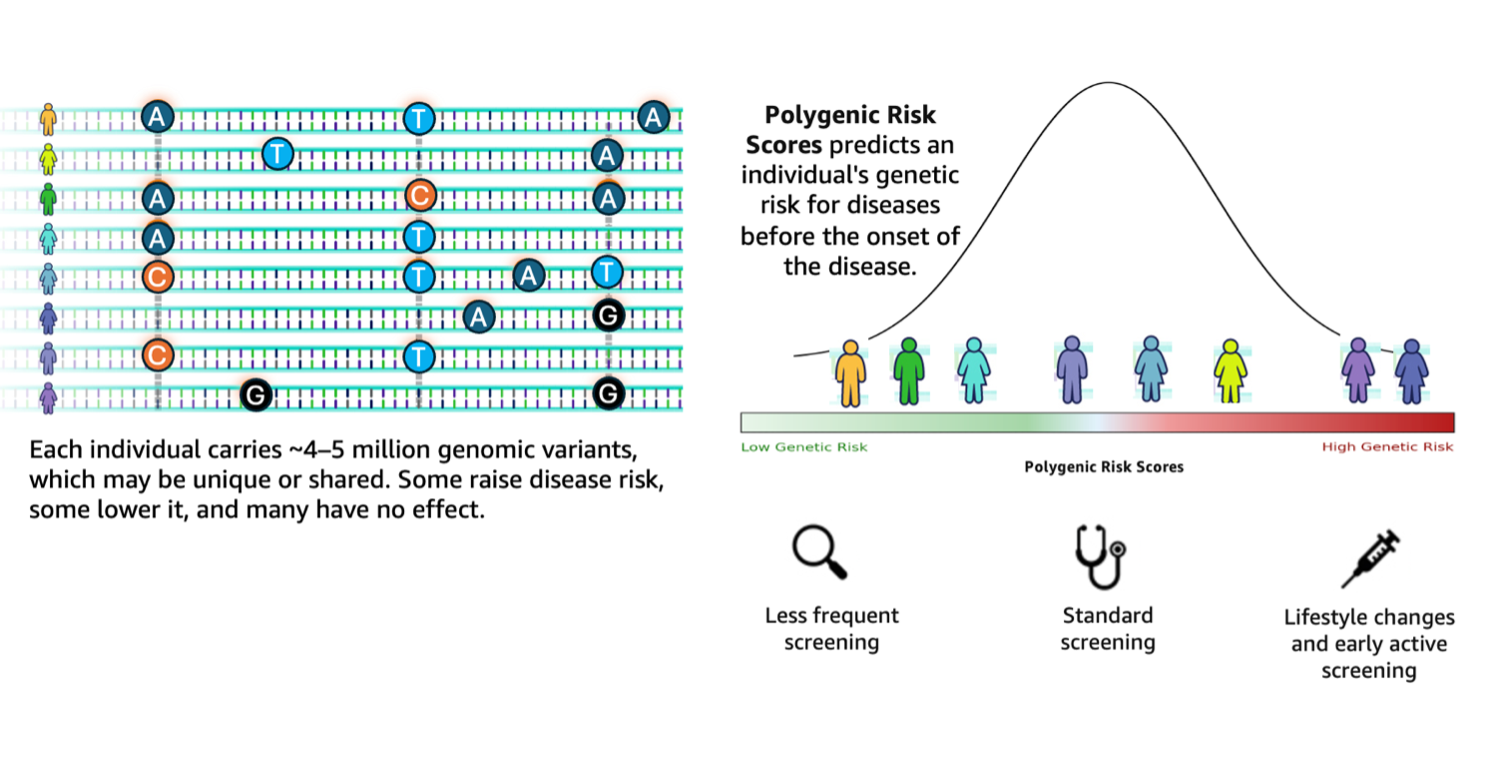
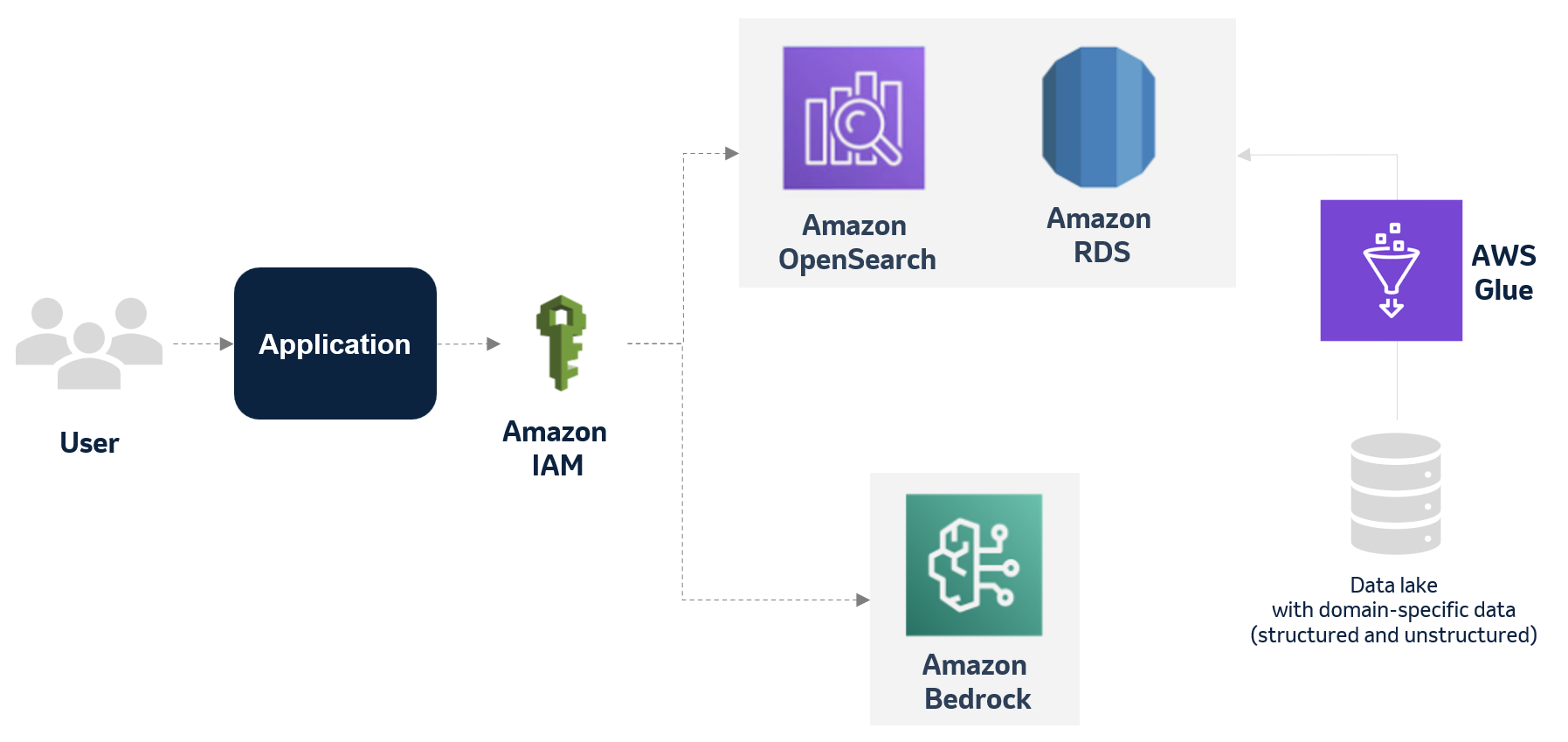
Accelerating genomics variant interpretation with AWS HealthOmics and Amazon Bedrock AgentCoreArtificial Intelligence In this blog post, we show you how agentic workflows can accelerate the processing and interpretation of genomics pipelines at scale with a natural language interface. We demonstrate a comprehensive genomic variant interpreter agent that combines automated data processing with intelligent analysis to address the entire workflow from raw VCF file ingestion to conversational query interfaces.
In this blog post, we show you how agentic workflows can accelerate the processing and interpretation of genomics pipelines at scale with a natural language interface. We demonstrate a comprehensive genomic variant interpreter agent that combines automated data processing with intelligent analysis to address the entire workflow from raw VCF file ingestion to conversational query interfaces. Read More
MSD explores applying generative Al to improve the deviation management process using AWS servicesArtificial Intelligence This blog post has explores how MSD is harnessing the power of generative AI and databases to optimize and transform its manufacturing deviation management process. By creating an accurate and multifaceted knowledge base of past events, deviations, and findings, the company aims to significantly reduce the time and effort required for each new case while maintaining the highest standards of quality and compliance.
This blog post has explores how MSD is harnessing the power of generative AI and databases to optimize and transform its manufacturing deviation management process. By creating an accurate and multifaceted knowledge base of past events, deviations, and findings, the company aims to significantly reduce the time and effort required for each new case while maintaining the highest standards of quality and compliance. Read More
Oligo Security has warned of ongoing attacks exploiting a two-year-old security flaw in the Ray open-source artificial intelligence (AI) framework to turn infected clusters with NVIDIA GPUs into a self-replicating cryptocurrency mining botnet. The activity, codenamed ShadowRay 2.0, is an evolution of a prior wave that was observed between September 2023 and March 2024. The […]
A unique take on the software update gambit has allowed “PlushDaemon” to evade attention as it mostly targets Chinese organizations. Read More
Editors from Dark Reading, Cybersecurity Dive, and TechTarget Search Security break down the depressing state of cybersecurity awareness campaigns and how organizations can overcome basic struggles with password hygiene and phishing attacks. Read More
A major spike in malicious scanning against Palo Alto Networks GlobalProtect portals has been detected, starting on November 14, 2025. […] Read More
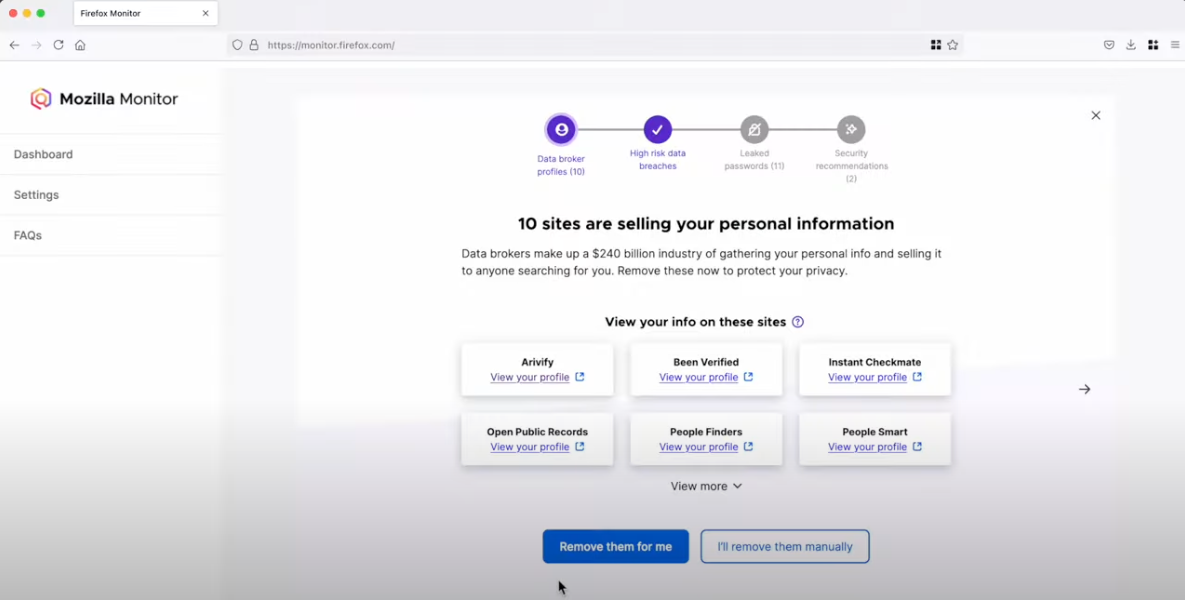
In March 2024, Mozilla said it was winding down its collaboration with Onerep — an identity protection service offered with the Firefox web browser that promises to remove users from hundreds of people-search sites — after KrebsOnSecurity revealed Onerep’s founder had created dozens of people-search services and was continuing to operate at least one of […]
Deep Learning-Based Regional White Matter Hyperintensity Mapping as a Robust Biomarker for Alzheimer’s Diseasecs.AI updates on arXiv.org arXiv:2511.14588v1 Announce Type: cross
Abstract: White matter hyperintensities (WMH) are key imaging markers in cognitive aging, Alzheimer’s disease (AD), and related dementias. Although automated methods for WMH segmentation have advanced, most provide only global lesion load and overlook their spatial distribution across distinct white matter regions. We propose a deep learning framework for robust WMH segmentation and localization, evaluated across public datasets and an independent Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. Our results show that the predicted lesion loads are in line with the reference WMH estimates, confirming the robustness to variations in lesion load, acquisition, and demographics. Beyond accurate segmentation, we quantify WMH load within anatomically defined regions and combine these measures with brain structure volumes to assess diagnostic value. Regional WMH volumes consistently outperform global lesion burden for disease classification, and integration with brain atrophy metrics further improves performance, reaching area under the curve (AUC) values up to 0.97. Several spatially distinct regions, particularly within anterior white matter tracts, are reproducibly associated with diagnostic status, indicating localized vulnerability in AD. These results highlight the added value of regional WMH quantification. Incorporating localized lesion metrics alongside atrophy markers may enhance early diagnosis and stratification in neurodegenerative disorders.
arXiv:2511.14588v1 Announce Type: cross
Abstract: White matter hyperintensities (WMH) are key imaging markers in cognitive aging, Alzheimer’s disease (AD), and related dementias. Although automated methods for WMH segmentation have advanced, most provide only global lesion load and overlook their spatial distribution across distinct white matter regions. We propose a deep learning framework for robust WMH segmentation and localization, evaluated across public datasets and an independent Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. Our results show that the predicted lesion loads are in line with the reference WMH estimates, confirming the robustness to variations in lesion load, acquisition, and demographics. Beyond accurate segmentation, we quantify WMH load within anatomically defined regions and combine these measures with brain structure volumes to assess diagnostic value. Regional WMH volumes consistently outperform global lesion burden for disease classification, and integration with brain atrophy metrics further improves performance, reaching area under the curve (AUC) values up to 0.97. Several spatially distinct regions, particularly within anterior white matter tracts, are reproducibly associated with diagnostic status, indicating localized vulnerability in AD. These results highlight the added value of regional WMH quantification. Incorporating localized lesion metrics alongside atrophy markers may enhance early diagnosis and stratification in neurodegenerative disorders. Read More
Near-Lossless Model Compression Enables Longer Context Inference in DNA Large Language Modelscs.AI updates on arXiv.org arXiv:2511.14694v1 Announce Type: cross
Abstract: Trained on massive cross-species DNA corpora, DNA large language models (LLMs) learn the fundamental “grammar” and evolutionary patterns of genomic sequences. This makes them powerful priors for DNA sequence modeling, particularly over long ranges. However, two major constraints hinder their use in practice: the quadratic computational cost of self-attention and the growing memory required for key-value (KV) caches during autoregressive decoding. These constraints force the use of heuristics such as fixed-window truncation or sliding windows, which compromise fidelity on ultra-long sequences by discarding distant information. We introduce FOCUS (Feature-Oriented Compression for Ultra-long Self-attention), a progressive context-compression module that can be plugged into pretrained DNA LLMs. FOCUS combines the established k-mer representation in genomics with learnable hierarchical compression: it inserts summary tokens at k-mer granularity and progressively compresses attention key and value activations across multiple Transformer layers, retaining only the summary KV states across windows while discarding ordinary-token KV. A shared-boundary windowing scheme yields a stationary cross-window interface that propagates long-range information with minimal loss. We validate FOCUS on an Evo-2-based DNA LLM fine-tuned on GRCh38 chromosome 1 with self-supervised training and randomized compression schedules to promote robustness across compression ratios. On held-out human chromosomes, FOCUS achieves near-lossless fidelity: compressing a 1 kb context into only 10 summary tokens (about 100x) shifts the average per-nucleotide probability by only about 0.0004. Compared to a baseline without compression, FOCUS reduces KV-cache memory and converts effective inference scaling from O(N^2) to near-linear O(N), enabling about 100x longer inference windows on commodity GPUs with near-lossless fidelity.
arXiv:2511.14694v1 Announce Type: cross
Abstract: Trained on massive cross-species DNA corpora, DNA large language models (LLMs) learn the fundamental “grammar” and evolutionary patterns of genomic sequences. This makes them powerful priors for DNA sequence modeling, particularly over long ranges. However, two major constraints hinder their use in practice: the quadratic computational cost of self-attention and the growing memory required for key-value (KV) caches during autoregressive decoding. These constraints force the use of heuristics such as fixed-window truncation or sliding windows, which compromise fidelity on ultra-long sequences by discarding distant information. We introduce FOCUS (Feature-Oriented Compression for Ultra-long Self-attention), a progressive context-compression module that can be plugged into pretrained DNA LLMs. FOCUS combines the established k-mer representation in genomics with learnable hierarchical compression: it inserts summary tokens at k-mer granularity and progressively compresses attention key and value activations across multiple Transformer layers, retaining only the summary KV states across windows while discarding ordinary-token KV. A shared-boundary windowing scheme yields a stationary cross-window interface that propagates long-range information with minimal loss. We validate FOCUS on an Evo-2-based DNA LLM fine-tuned on GRCh38 chromosome 1 with self-supervised training and randomized compression schedules to promote robustness across compression ratios. On held-out human chromosomes, FOCUS achieves near-lossless fidelity: compressing a 1 kb context into only 10 summary tokens (about 100x) shifts the average per-nucleotide probability by only about 0.0004. Compared to a baseline without compression, FOCUS reduces KV-cache memory and converts effective inference scaling from O(N^2) to near-linear O(N), enabling about 100x longer inference windows on commodity GPUs with near-lossless fidelity. Read More